Add arc labels to plotted phylogeny
arc_labels(phy, tips, ...)
# S3 method for default
arc_labels(
phy = NULL,
tips,
text,
plot_singletons = TRUE,
ln.offset = 1.02,
lab.offset = 1.06,
cex = 1,
orientation = "horizontal",
...
)Arguments
- phy
An object of class phylo.
- tips
A character vector (or a list) with names of the tips that belong to the clade or group. If multiple groups are to be plotted, tips must be given in the form of a list.
- ...
Further arguments passed to or from other methods.
- text
Desired clade label.
- plot_singletons
Logical. If TRUE (default), adds arcs (and labels) to single tip lineages. If FALSE, no arc or labels will be plotted over that tip..
- ln.offset
Line offset (as a function of total tree height)
- lab.offset
Label offset.
- cex
Character expansion
- orientation
Orientation of the text. Can be “vertical”, “horizontal”, or “curved”.
Value
NULL
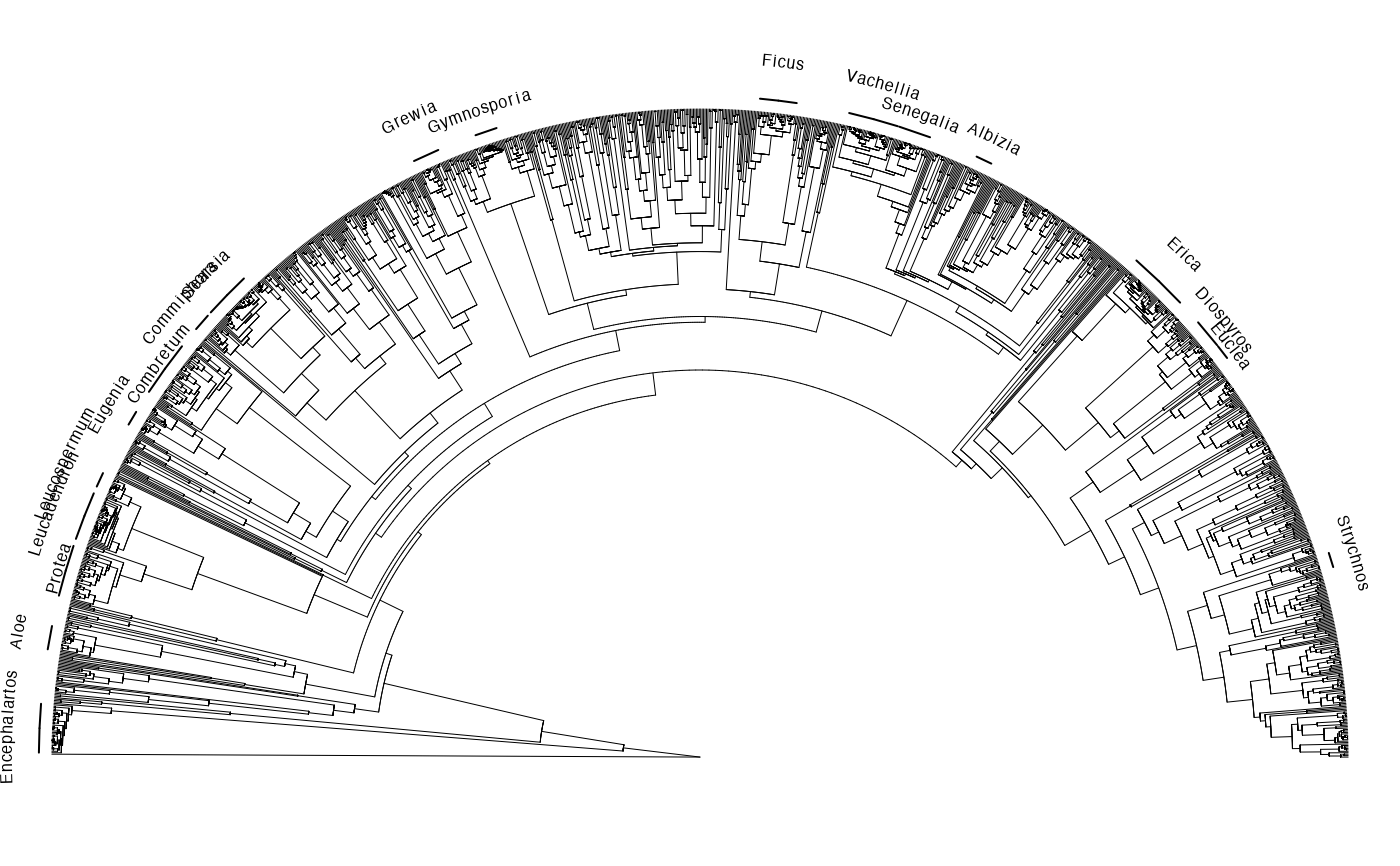
Examples
# \donttest{
old.par <- par(no.readonly = TRUE)
require(ape)
data(africa)
par(mai=rep(0,4))
plot(africa$phylo, type = "fan", show.tip.label=FALSE,
open.angle = 180, edge.width=0.5)
y <- data.frame(species=africa$phylo$tip.label)
y$genus <- gsub("_.*", "\\1", y$species)
fx <- split(y, f=y$genus)
suppressWarnings(invisible(lapply(fx, function(x) {
y <- seq(from = 1.03, to = 1.09, by = ((1.09 - 1.03)/(length(fx) - 1)))
z <- sample(y, 1, replace = FALSE, prob = NULL)
if(nrow(x) > 10L) arc_labels(phy = africa$phylo, tips=x$species,
text=as.character(unique(x$genus)),
orientation = "curved", cex=0.5,
lab.offset = z)
})))
 par(old.par)
# }
par(old.par)
# }